Immunomics Services
Heavy Chain of BCR Sequencing Service
B cells are an essential part of the adaptive immune system. The diversity of heavy chains, particularly the CDR3 structural domain of VH in heavy chains, is sufficient to allow identical IgM molecules to distinguish between various semi-antigens and protein antigens. Based on years of experience in immunomics, CD Genomics offers the heavy chain of BCR sequencing services to advance your immunomics projects.
Introduction to Heavy Chain of BCR
B cell-mediated immunity is dependent on Ig Abs produced by the clonal expansion of B cells. BCR is the membrane-bound form of Ab and consists of two heavy chains (H) and two light chains (L) paired in a heterodimeric fashion. Each chain contains a variable (V) region, and the V regions of the H and L chains together constitute the Ag binding site.
In humans, heavy chains are formed by recombination of one of 38 to 46 functionally variable (VH) genes with one of 23 diversity (DH) genes and one of 6 linkages (JH) genes. The recombination process also introduces diversity at the junctions of the VH, DH, and JH genes by removing and adding nucleotides. The CDR3 region of the BCR includes light chain V and J gene fragments and heavy chain V, D, and J gene fragments.
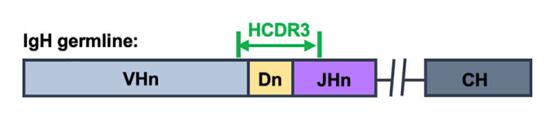 Fig.1 A representation of the coding region of an IgH heavy-chain variable region is presented. (Ou, T., et al., 2022)
Fig.1 A representation of the coding region of an IgH heavy-chain variable region is presented. (Ou, T., et al., 2022)
Rationale of the Heavy Chain Sequencing and Analysis
Adaptive immune complex receptor sequencing allows high-throughput analysis of different BCR complexes by full-length V(D)J sequencing. Cells sharing the same IGHV and IGHJ genes have sufficient similarity in their H-chain linkage sequences based on fixation or adaptation to be clustered as clones. Therefore, although antibodies are encoded by sequences of both light and heavy chains, analysis of heavy chains is considered sufficient to determine immunome characteristics such as vaccination response. Studies based on the heavy chain of BCR repertoire are now available, for example, for mechanistic studies in cancer and infectious diseases.

Research on Tumor
A large-scale assembly and analysis of a pan-cancerous species tumor-infiltrating BCR repertoire reveal the prevalence of somatic copy number variants in the MICA and MICB genes associated with antibody-dependent cell-mediated cytotoxicity in tumors with elevated B-cell activity, providing a potential resource for the future development of cancer immunotherapies.

Research on Infectious Diseases
In addition, the construction of the heavy chain of BCR repertoire, analysis of somatic hypermutations of different immunoglobulins, CDR3 length, correlation of Ig heavy chain features with T cell subsets, clonotypes, and clonal expansion analysis provide key information for the development of HIV neutralizing antibodies.
Our Services
Next-generation sequencing technologies are increasingly being used to query antibody or B-cell receptor (BCR) repertoires and sequence libraries, creating the opportunity to learn more about our immune system. Heavy chains determine different antibody types, and increasingly BCR libraries containing full-length sequences are being generated that focus primarily on heavy chain sequences.
CD Genomics has many years of experience in immunomics research and can provide efficient and reliable heavy chain of BCR repertoire sequencing services to our clients.
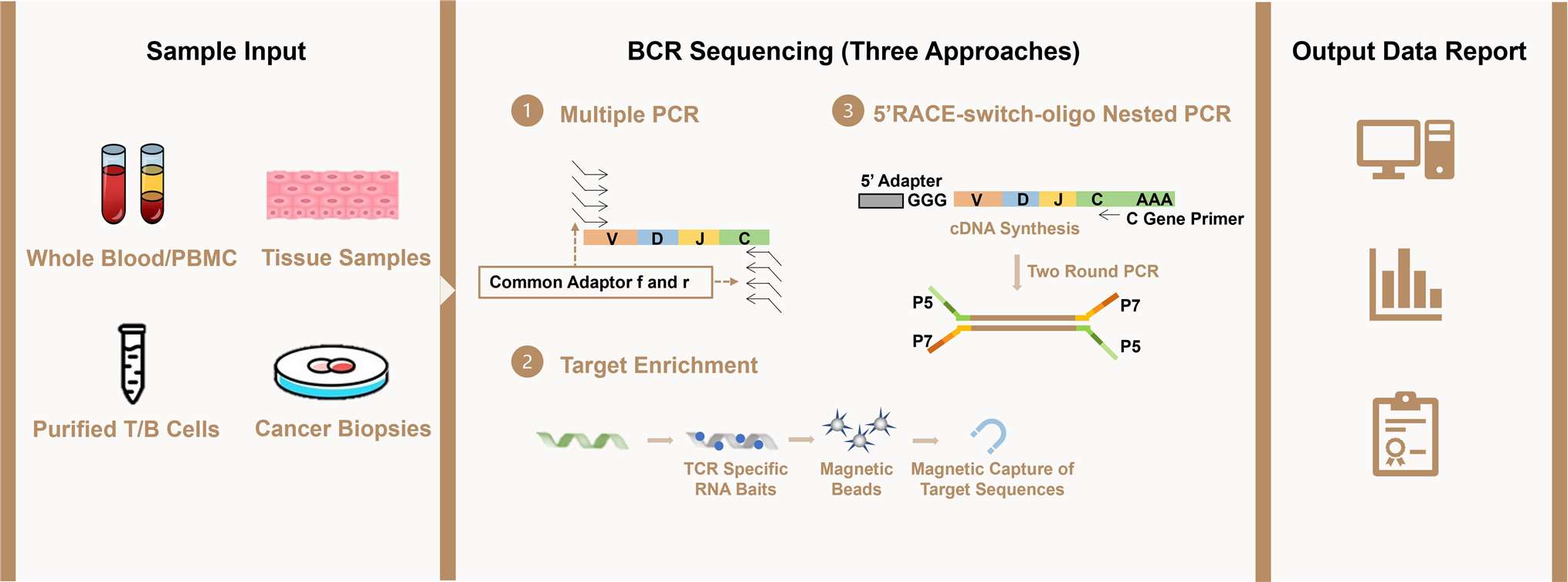
Workflow of Heavy Chain of BCR Sequencing
Why Choose Us
| Multiple Library Construction Methods. | |
| Advanced High-throughput Sequencing Platform. | |
| Professional Data Analysis Team. |
As a professional immunomics research platform, CD Genomics is dedicated to providing important information on the immune response of the body through sequencing and analysis of the BCR repertoire. Please contact us for more information.
Reference
- Ou, T., He, W., Quinlan, B. D., Guo, Y., Tran, M. H., Karunadharma, P., ... & Farzan, M. (2022). Reprogramming of the heavy-chain CDR3 regions of a human antibody repertoire. Molecular Therapy, 30(1), 184-197.
