Immunomics Services
Immunomics Bioinformatics Services
Immunomics studies require large cohorts of sample sizes. With data production and quality control finished in-depth downstream analysis of the data can begin. CD Genomics' bioinformatics analysis for immunomics keeps pace with new bioinformatics tools, offering clients endless possibilities for analyzing their data.
Introduction to Bioinformatics
Bioinformatics helps to interpret large-scale and high-dimensional data in immunomics. The first analysis step consists of data exploration using unbiased and unsupervised methods, such as hierarchical clustering or principal component analysis. Following this, hypothesis testing is performed using statistical methods to compare gene expression levels of different groups in the study cohort, providing a primary readout.
Then comes the search for biological explanations of the data model, including potential signaling pathway changes, chromatin region status, and potential ligand-receptor interactions between different cell types. And then is to validate the obtained data models in the human dataset.
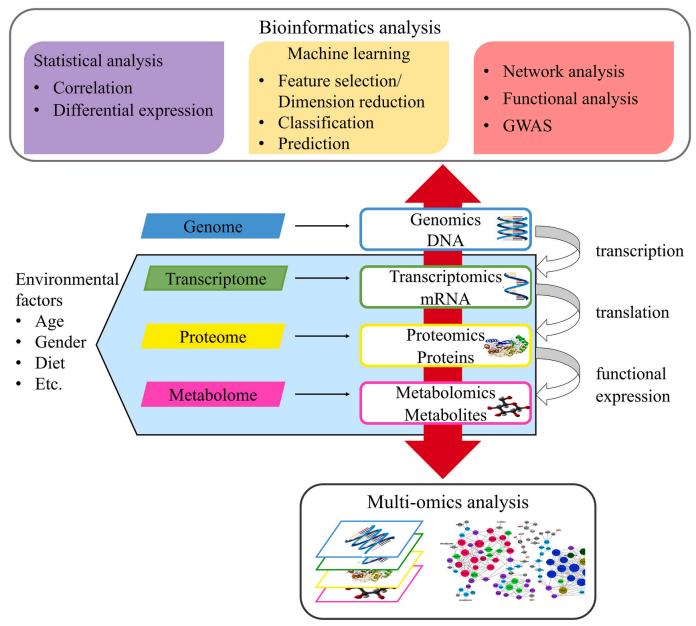 Fig.1 Bioinformatics analysis for omics data.
(Tan, M. S., et al., 2021)
Fig.1 Bioinformatics analysis for omics data.
(Tan, M. S., et al., 2021)
Applications of Bioinformatics for Immunomics
The wealth of omics data and powerful algorithms facilitate a better understanding of the complex cellular and molecular interactions operating within the immune system, including a comprehensive description of immune homeostasis and immune variation across the population, molecular definition of cell types and states, dynamic descriptions of local and systemic immune responses, and the interaction of the immune system within organ systems.
Our Services
We support hypothesis-driven research that combines omics data with computational modeling and experimental validation as a powerful method for data generation, interpretation, and effective knowledge acquisition.
CD Genomics is made up of bioinformaticians, data scientists, and biologists. We can provide DNA sequencing data analysis, RNA sequencing data analysis, single-cell sequencing data analysis, epigenomic data analysis, and proteomic and metabolomic data analysis. For each project, we assemble an optimal service team to develop new analysis modules, perform statistical analysis, and draw beautiful pictures for the final paper for you.

Discovering Cell Types
We analyze RNA sequencing, single-cell RNA sequencing, and single-cell NGS data to identify, quantify, and compare immune cell types at different developmental stages, niches, and other conditions, to describe the intracellular pathways by which immune cells respond to stimuli, and to determine the interactions between immune system cells and between immune cells and other tissues.

Studying Tumor Immune Evasion
In cancer research, RNA sequencing of tumors enables the characterization of their immune microenvironment. Identification of tumor-infiltrating immune cells helps to study the mechanisms of immune evasion and therapeutic opportunities. Tumor RNA or DNA sequencing data can also be used to identify potential new antigens for cancer vaccine development.

Uncovering Host-Pathogen Interactions
Pathogenic proteins cause extensive remodeling of host immune cells. RNA sequencing of immune cells from pathogen infection models in vivo or in vitro can describe the mechanisms by which various cell types of the immune system respond to antigens, rely on information, and neutralize pathogens.

Characterizing Immune Complexes
More targeted immune sequencing approaches can be used to study complexes of lymphocyte receptors under different conditions or before and after infection or vaccination. Such data are often generated in conjunction with single-cell RNA-seq, allowing gene expression to be correlated with clonal properties.
Why Choose Us
Our Immunomics Bioinformatics Service is designed for scientists who want easy access to the right bioinformatics expertise. With our technical team, we help our clients worldwide to find the best solution for their research needs. Please contact us for more information.
Reference
- Tan, M. S., Cheah, P. L., Chin, A. V., Looi, L. M., & Chang, S. W. (2021). A review on omics-based biomarkers discovery for Alzheimer's disease from the bioinformatics perspectives: statistical approach vs machine learning approach. Computers in biology and medicine, 139, 104947.
