Immunomics Services
BCR Repertoire Analysis Services
The diversity of B-cell receptors (BCR) is one of the cores that make up the complex immune system and serves as a key defense component to protect the body from invasion by viruses, bacteria, and other pathogens. CD Genomics provides customized services for BCR sequencing services to global clients, providing reliable data for immunomics projects.
Introduction to BCR Repertoire
B cells play an important role in the pathogenesis of many, perhaps all, immune-mediated diseases. Each B cell expresses a BCR, and the various BCRs expressed by the total B cell population of an individual is referred to as the BCR repertoire. Further diversification occurs after the B cell responds to the antigen.
Diversity in the B Cell Repertoire
The formation and revision of BCR is a highly dynamic process. The number of lymphocyte clones per lymphocyte is highly variable, depending on the cell specificity and history of antigen exposure.
- Diversity in the Naive B Cell Repertoire
The genes encoding the IgH and IgL chains are assembled from IGH variable (V), diversity (D), and linkage (J) and IGK or IGL V and J gene fragments and a small number of C region fragments during the early stages of B cell development in the bone marrow. The combinatorial diversity generated by V(D)J recombination is further increased by the imprecision of the rearrangement process, which deletes and adds nucleotides while linking Ig gene fragments.
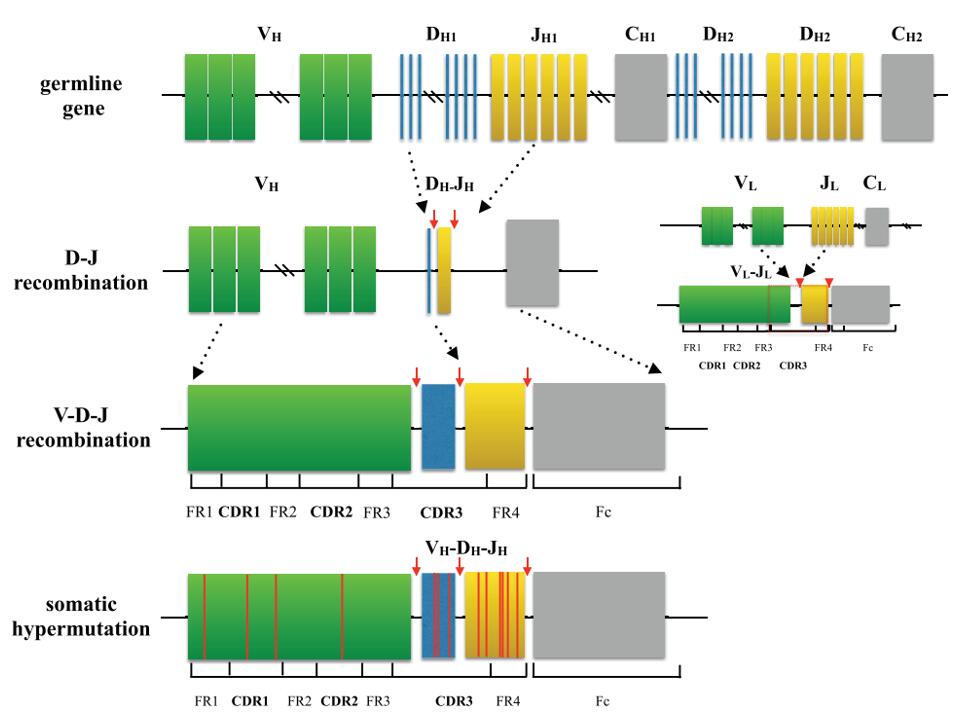 Fig.1 Process of generating a diverse BCR repertoire. (Hou, D., et al., 2016)
Fig.1 Process of generating a diverse BCR repertoire. (Hou, D., et al., 2016)
- Diversity in the Antigen-experienced B Cell Repertoire
Antigen exposure clonally selects and activates reactive B cells, inducing them to proliferate and differentiate into antibody-secreting effector cells as well as memory cells that can generate an effective response upon antigen re-challenge. Multivalent antigens can activate B cells by direct BCR cross-linking, while the monovalent protein antigens require the help of T cells. - Diversity at the Cellular Level
The CD19+ B cell populations contained in peripheral blood samples are mainly antigen-inexperienced (naive) and antigen-experienced B cells. Due to the lack of clonal amplification, the complex of immature and mature naive B cells is highly diverse compared to the antigen-experienced B cell compartment. The diversity of cellular B cells is not limited to blood. Similar levels of diversity are found in lymphoid and even non-lymphoid organs and tissues.
Our Services
Our understanding and definition of the BCR repertoire in the context of disease reveals a complex B cell architecture that could provide new insights into pathogenesis and therapy. As a professional immunomics research platform, CD Genomics provides high-quality BCR repertoire sequencing and analysis services to provide reliable data for your projects. We provide both the heavy chain and light chain of the BCR Sequencing Service.
- Comparison of BCR Repertoire in Defined Cell Compartments
Flow cytometric classification allows the classification of B cell subpopulations based on the expression of B cell surface markers for bulk or single cell analysis. Based on the same markers, CD Genomics offers direct comparison services of identified cellular compartments in individual studies or across studies - Data Analysis Services
CD Genomics has a professional bioinformatics team. After obtaining sequencing information from BCR libraries, we provide efficient and comprehensive data processing and analysis services to our clients.
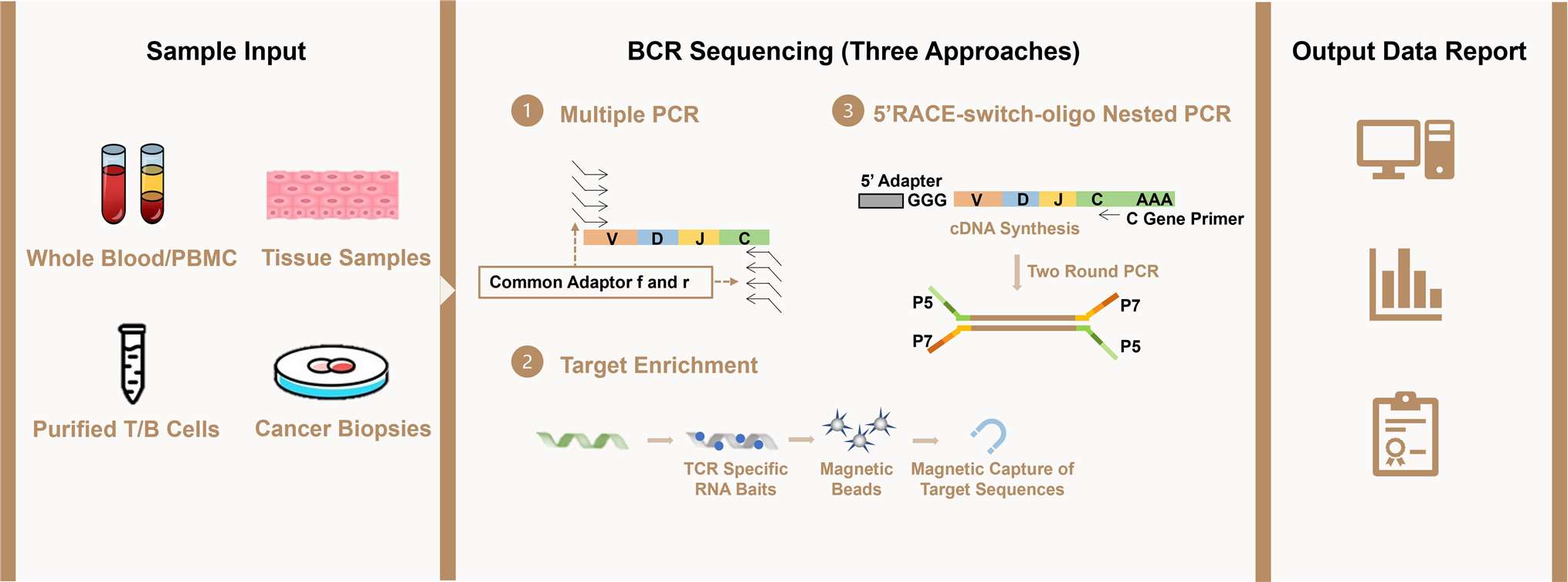
Workflow of BCR Repertoire Analysis
Why Choose Us
Based on years of experience in immunomics research, CD Genomics provides efficient and reliable BCR repertoire analysis to clients worldwide to explore effective vaccines by assessing dynamic changes in the immune pool following antigen stimulation and disease diagnosis through BCR repertoire analysis. Please contact us for more information.
| Multiple Library Construction Methods. | |
| Advanced High-throughput Sequencing Platform. | |
| Professional Data Analysis Team. |
Reference
- Hou, D., Chen, C., Seely, E. J., Chen, S., & Song, Y. (2016). High-throughput sequencing-based immune repertoire study during infectious disease. Frontiers in immunology, 7, 336.
