Immunomics Services
TCR Repertoire Analysis Services
T cell-mediated antigen recognition depends on the interaction of the T cell receptor (TCR) with the antigen-major histocompatibility complex (MHC) molecule. Analysis of the TCR repertoire may contribute to a better understanding of immune system characteristics and the etiology and progression of diseases, particularly those with unknown antigenic triggers. With many years of research experience in immunomics, CD Genomics offer our clients professional, efficient, and full-coverage services for the analysis of TCR repertoire, including the α, β, γ, and δ chains of T cells.
What Is TCR?
The TCR is a highly diverse heterodimer, consisting of a combination of alpha and beta chains expressed by most T cells, or gamma and delta chains expressed by T cells in the peripheral blood (1-5%) and by T cells in mucosal sites. Similar to the immunoglobulins expressed by B cells, the TCR chain consists of a variable region, which is important for antigen recognition, and a constant region. The variable regions of the TCR α and δ chains are encoded by several variable (V) and linkage (J) genes, while the TCR β and γ chains are additionally encoded by diversity (D) genes.
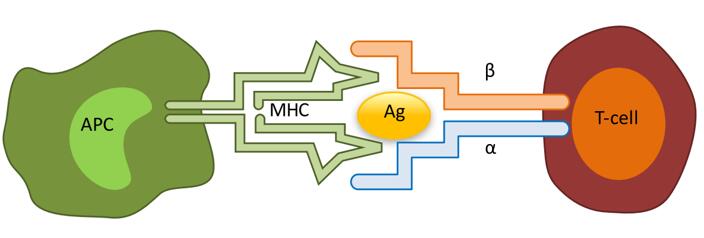 Fig.1 Interaction between the MHC and the αβ TCR. (Rosati, E., et al., 2017)
Fig.1 Interaction between the MHC and the αβ TCR. (Rosati, E., et al., 2017)
During VDJ recombination, a random allele of each gene segment recombines with other gene segments to form a functional variable region. The recombination of the variable region with the constant gene segment generates a functional TCR strand transcript.
Important Roles of TCR Repertoire
TCR rearrangements can vary greatly with the onset and progression of the disease, which is why scientists are increasingly interested in determining the status of immune rearrangements in different disease conditions, such as cancer, autoimmune diseases, inflammatory diseases, and infectious diseases.
For example, TCR rearrangement studies can be used to analyze the effects of autologous stem cell transplantation on the T-cell population of patients with multiple sclerosis. In addition, as cytotoxic T cells can kill tumor cells in cancer upon recognition of tumor-specific antigens, TCR repertoire analysis can be used to study the redistribution of tumor-infiltrating lymphocytes and to identify specific T cell clonotypes involved in this process.
Our Services
The sequencing and analysis of TCR libraries is a meaningful direction to enhance the understanding of the immune system, especially for mechanistic studies of infections, autoimmune diseases, and cancer, and to help discover new therapeutic agents.
- Selectable Starting Materials
One of the most basic but important decisions a scientist should make when choosing a TCR analysis method is the starting material, which is to use genomic DNA (gDNA) or RNA to conduct TCR sequencing. Both types of starting materials have advantages and disadvantages. CD Genomics can offer both types of TCR sequencing services for our clients. - Sequencing and Analysis of Different Chains of T Cells
As we all know, the TCR consists of variable regions and constant regions. The TCR repertoire in humans includes four types, which are α chain, β chain, γ chain, and δ chain. CD Genomics provides sequencing and analysis services for these four types of TCR repertoire.
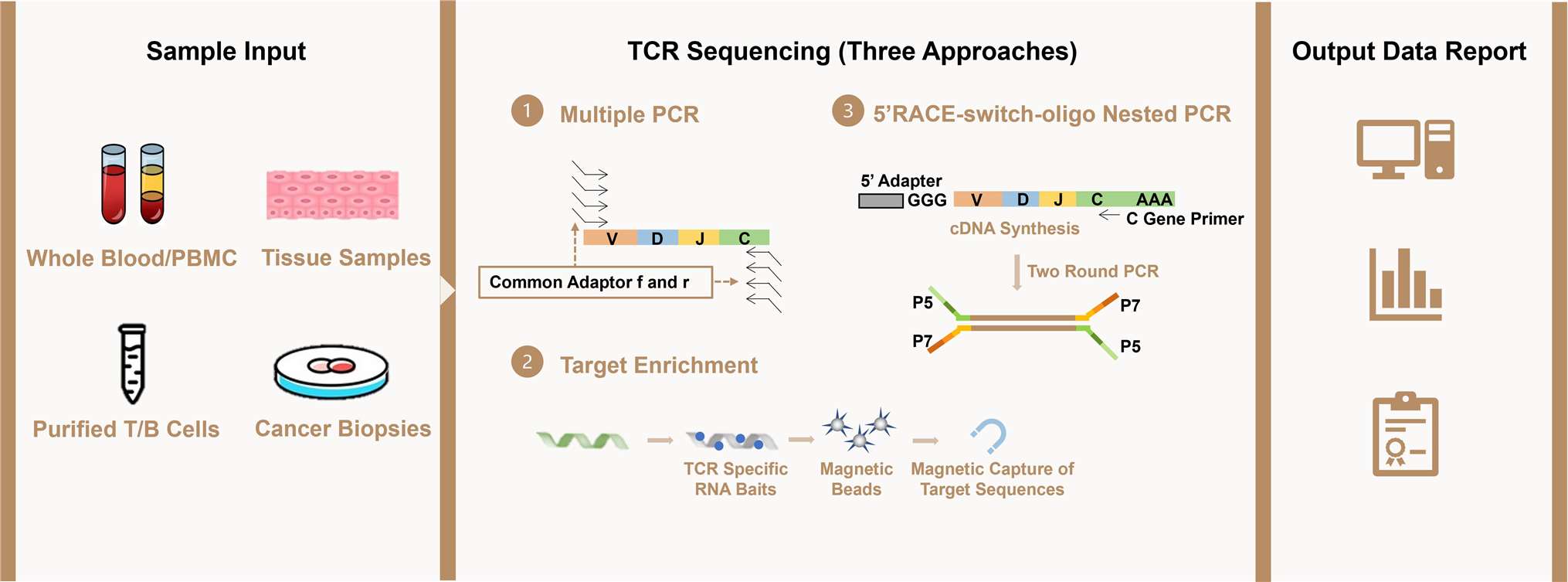
Workflow of TCR Repertoire Analysis
Why Choose Us
| Multiple Library Construction Methods. | |
| Advanced High-throughput Sequencing Platform. | |
| Professional Data Analysis Team. |
CD Genomics has specialized in immunomics research for many years and has provided professional analysis of the TCR repertoire for a number of research institutions. Our company is committed to advancing the study of multiple disease mechanisms and the development of novel therapies through immunomics research. Please contact us for more information.
Reference
- Rosati, E., Dowds, C. M., Liaskou, E., Henriksen, E. K. K., Karlsen, T. H., & Franke, A. (2017). Overview of methodologies for T-cell receptor repertoire analysis. BMC biotechnology, 17(1), 1-16.
