Immunomics in Autoimmune Diseases
The human immune system is a multifaceted network of functionally diverse cells that express a wide range of receptors that work together to respond to infection, eliminate precancerous cells, and maintain metabolic health. Small errors in this delicate immune response can cause a range of autoimmune diseases.
Introduction to Autoimmune Diseases
More than 80 autoimmune diseases have been described. They can be systemic, such as systemic lupus erythematosus, which can affect the skin, joints, kidneys, and central nervous system. They can also be organ-specific, such as type 1 diabetes, which mainly affects the pancreas. Loss of B-cell or T-cell tolerance is often associated with autoimmunity. Studying human autoimmune disease at multiple levels through immunomics helps us to understand its mechanisms and drivers.
TCR/BCR Sequencing in Autoimmune Diseases
The immune repertoire (IR) is the sum of the functionally diverse B and T lymphocytes in an individual's circulatory system at any given time. T and B cells mediate the body's cellular and humoral immune responses and recognize and bind antigens through their respective surface T-cell receptors (TCRs) and B-cell receptors (BCRs) to clear pathogens or tumor cells from the body. Sequencing IR from patients can help to unravel pathogenesis and provide potential therapeutic targets.
For example, by sequencing single-cell T-cell receptor (TCR)/B-cell receptor (BCR) in patients with systemic lupus erythematosus (SLE) and normal controls (NC), it is found that SLE patients have increased TCR and BCR clonotypes and that SLE patients exhibit a bias towards the use of TCR and BCR V(D)J genes. The most used V gene in TCRα is TRAV40 in NC and TRAV16 in SLE and the most used V gene in BCR IGH is IGHV3-21 in NC and IGHV3-23 in SLE.
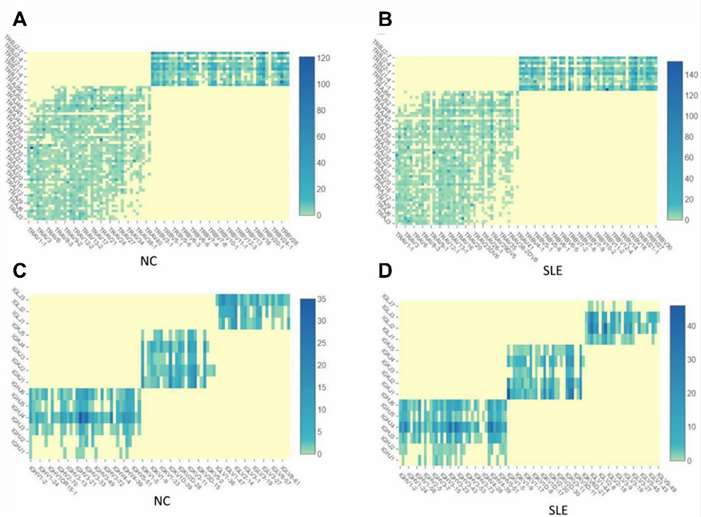 Fig.1 The TCR and BCR clonotypes are increased in SLE than in NC. (Zheng, F., et al., 2021)
Fig.1 The TCR and BCR clonotypes are increased in SLE than in NC. (Zheng, F., et al., 2021)
Epigenomics in Autoimmune Diseases
Genes seem to explain only a small part of the heritability of some autoimmune diseases. Epigenetic markers may explain part of the heritability of the deletion. The possibility of elucidating the mechanisms by which gene expression may be altered has therefore become an area of intense research in recent years, including the identification of epigenetic modifications of gene regulatory regions.
Transcriptomics in Autoimmune Diseases
Another approach to studying autoimmune diseases is transcriptomics, the study of gene expression levels in disease, particularly using microarrays, but more recently using next-generation RNA sequencing (RNA-Seq). Transcriptomic studies provide powerful tools to demonstrate the heterogeneity of tissue cells involved in various immune-inflammatory conditions, identify pathogenic cell populations, and reveal mechanisms of disease development.
Proteomics in Autoimmune Diseases
Proteomics is a high-throughput analysis of the expression, function, and interactions of proteins expressed in cells, tissues, or organisms. Protein arrays and other multiplex screening technologies are powerful tools for studying the pathophysiology and specificity of autoimmune responses. It may become possible to identify the antigenic fingerprint of each patient. This may be a critical step in targeting major disease-causing antigens.
A study using a proteomic approach to discover biomarkers for rheumatoid arthritis, using mass spectrometry for serum protein profiling, identified serum amyloid A4 and vitamin D binding protein as potential biomarkers that may be associated with the inflammatory response and joint destruction that accompany rheumatoid arthritis.
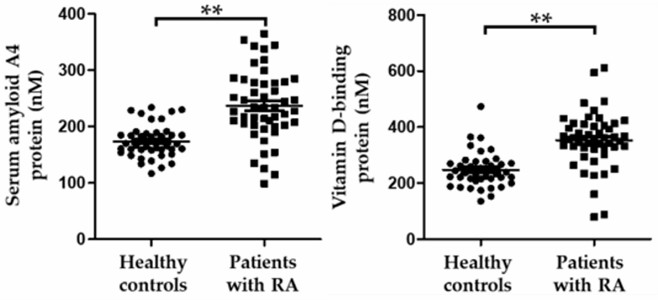 Fig.2 Dot plots of selected biomarker candidates in healthy controls and patients with rheumatoid arthritis. (Mun, S., et al., 2019)
Fig.2 Dot plots of selected biomarker candidates in healthy controls and patients with rheumatoid arthritis. (Mun, S., et al., 2019)
CD Genomics has been focusing on immunomics for many years. We offer a wide range of immunomics-related services, including immunogenomics analysis, immune cell epigenomics analysis, immune cell transcriptomics analysis, immune cell proteomics analysis, single-cell omics analysis for immune cells, and immunomics bioinformatics services to help clients understand the mechanisms of the immune system and discover novel drug and vaccines.
References
- Zheng, F., Xu, H., Zhang, C., Hong, X., Liu, D., Tang, D., ... & Dai, Y. (2021). Immune cell and TCR/BCR repertoire profiling in systemic lupus erythematosus patients by single-cell sequencing. Aging (Albany NY), 13(21), 24432.
- Mun, S., Lee, J., Park, A., Kim, H. J., Lee, Y. J., Son, H., ... & Kang, H. G. (2019). Proteomics approach for the discovery of rheumatoid arthritis biomarkers using mass spectrometry. International journal of molecular sciences, 20(18), 4368.
