Immunomics in Transplantation
Organ transplantation has evolved from an experimental approach in the 20th century to an established and practical option for the ultimate treatment of patients with end-organ dysfunction. Immunomics studies have greatly influenced transplantation research and highlighted their potential for a better understanding of graft outcomes.
Introduction to Transplantation
Organ transplantation is the first choice of treatment for end-stage organ failure. However, all transplant patients are at risk of complications, including allograft rejection and death. Over the past few decades, organ transplantation has evolved rapidly as various solid organs, such as the liver, kidney, pancreas, heart, and lung, and hematopoietic cell transplantation (HCT). Histological analysis of graft biopsies remains the gold standard for assessing allograft injury, but it is an invasive procedure that is prone to sampling errors. Over the past decade, there has been an increasing effort to develop minimally invasive procedures for monitoring allograft injury.
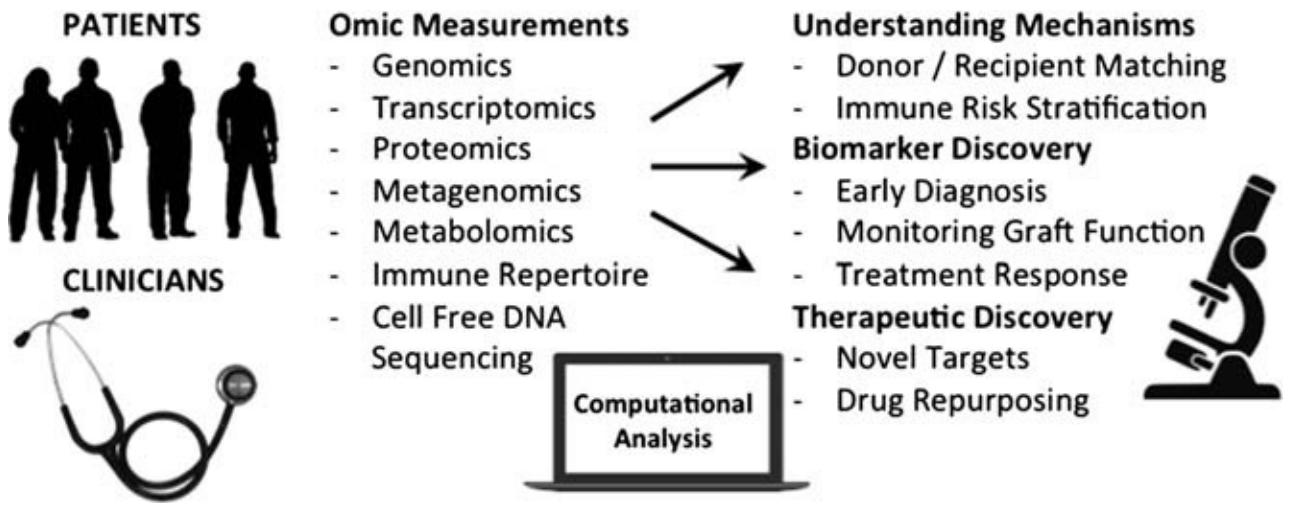 Fig.1 Workflow of immunomics studies for organ transplantation. (Sirota, M., & Sarwal, M. M., 2017)
Fig.1 Workflow of immunomics studies for organ transplantation. (Sirota, M., & Sarwal, M. M., 2017)
TCR/BCR Sequencing in Transplantation
It is thought that the crux of premature organ failure is primarily due to humoral damage caused by donor-specific B-cell responses and allograft rejection by alloreactive T cells. Immune complexes are the sum of functionally distinct B and T cells in a person's circulation at any given moment. The repertoire is complex and dynamic. The complexity of diversity arises from processes such as the combinatorial nature of V(D)J rearrangements, linkage diversity, pairing of heavy and light chains, and somatic hypermutation. The recent advent of high-throughput sequencing technologies has enabled researchers to sample and study the immune repertoire in blood and other tissues on a large scale.
Transcriptomics in Transplantation
Transcriptomes provide new insights into complex biological systems. Researchers are increasingly using the transcriptome to comprehensively characterize complex organs in health and disease. The diversity of immune cell types in transplanted organs undergoing rejection, some present at low frequencies, makes the transcriptome well suited to characterizing transplantation pathologies as it can quantify subtle transcriptional differences between rare cell types.
Epigenomics in Transplantation
The development of high-throughput microarrays has made Epigenome-Wide Association Analysis (EWAS) possible, recently enabling interrogation of the entire epigenome at approximately 850,000 loci. EWAS analysis allows assessment of the recipient's graft outcome, the apparent molecular and metabolic alterations that occur within the recipient as a result of multiple factors, such as the introduction of a foreign donor genome, and the recipient's response to immunosuppressive drugs.
Proteomics in Transplantation
A variety of biofluids have been successfully tested for the presence of potential proteomic biomarkers, these include serum, plasma, urine, and whole blood. Solid organ transplantation studies have provided important evidence for the potential of proteomics-based biomarkers in acute and chronic renal rejection, delayed graft function, and early detection of allograft health decline.
In a study based on 219 adult heart transplant recipients, pre-transplant serum samples are analyzed, and pre-transplant circulating levels of CLEC4C, a protein marker for plasmacytoid dendritic cells (pDC), are found to identify heart transplantation receptors at risk of primary graft dysfunction (PGD). That is, those patients with peripheral blood mononuclear cell populations enriched in pDCs may be at higher risk for transient interferon and TNF-mediated cardiotoxicity due to activation of pDCs by oxidized mtDNA released during ischemic and/or reperfusion injury of the cardiac allograft.
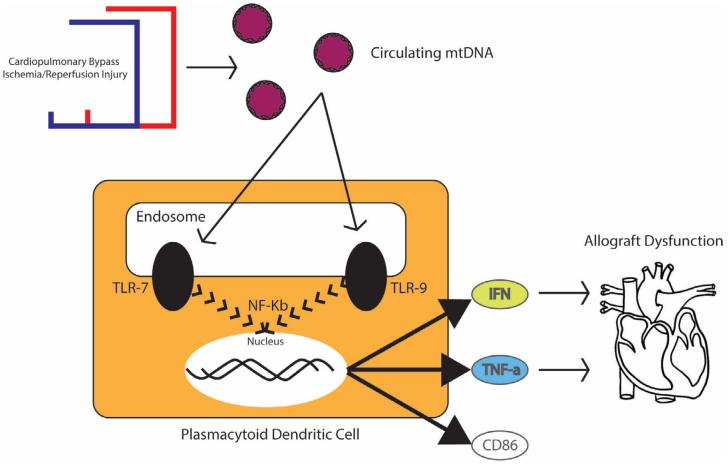 Fig.2 Mechanisms and roles of the innate immune system and pDCs in the pathogenesis of PGD. (Truby, L. K., et al., 2021)
Fig.2 Mechanisms and roles of the innate immune system and pDCs in the pathogenesis of PGD. (Truby, L. K., et al., 2021)
CD Genomics has been involved in immunomics projects for a number of companies around the world, helping clients to resolve bottlenecks in their projects. We offer a wide range of immunomics-related services, including immunogenomics analysis, immune cell epigenomics analysis, immune cell transcriptomics analysis, immune cell proteomics analysis, single-cell omics analysis for immune cells, and immunomics bioinformatics services to help clients understand the mechanisms of the immune system and discover novel drug and vaccines.
References
- Sirota, M., & Sarwal, M. M. (2017). Transplantomics: toward precision medicine in transplantation research. Transplantation, 101(8), 1777-1782.
- Truby, L. K., Kwee, L. C., Agarwal, R., Grass, E., DeVore, A. D., Patel, C. B., ... & Holley, C. L. (2021). Proteomic profiling identifies CLEC4C expression as a novel biomarker of primary graft dysfunction after heart transplantation. The Journal of Heart and Lung Transplantation, 40(12), 1589-1598.
